FAQ/Help
FAQs
What is CPRiL?
What is the benchmark dataset?
How are the entities annotated?
Which method is used for classification?
CPRiL is a webserver for exploring functional compound-protein relationships that are extracted automatically from PubMed literature. The functional relationships include directly interacting, up- or down-regulating each other (directly or indirectly), part of each other, or as cofactors of the related proteins.
The benchmark dataset has been manually curated. This dataset consists of 5,562 compound-protein pairs and 2,613 sentences extracted from PubMed abstracts. This dataset was used as a training dataset of three different machine learning approaches. More details of the benchmark can be found in the related manuscript: here).
The entities (compounds and proteins) are annotated by using NCBI PubTator Central(PTC).
For the classification, CPRiL is using Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT).
Searches
CPRiL offers different types of searching to receive functional compound-protein relationships:
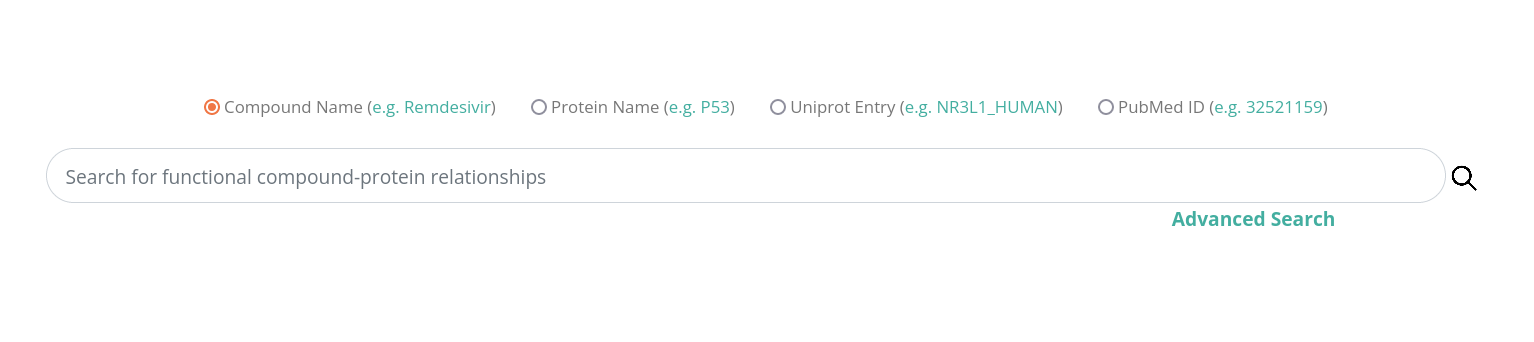
Compound Name
You can search with a specific compound name and get the best match of the given name, next you can select one of the suggested compound names.
The result list shows all proteins that have functional relationship to the selected compound.
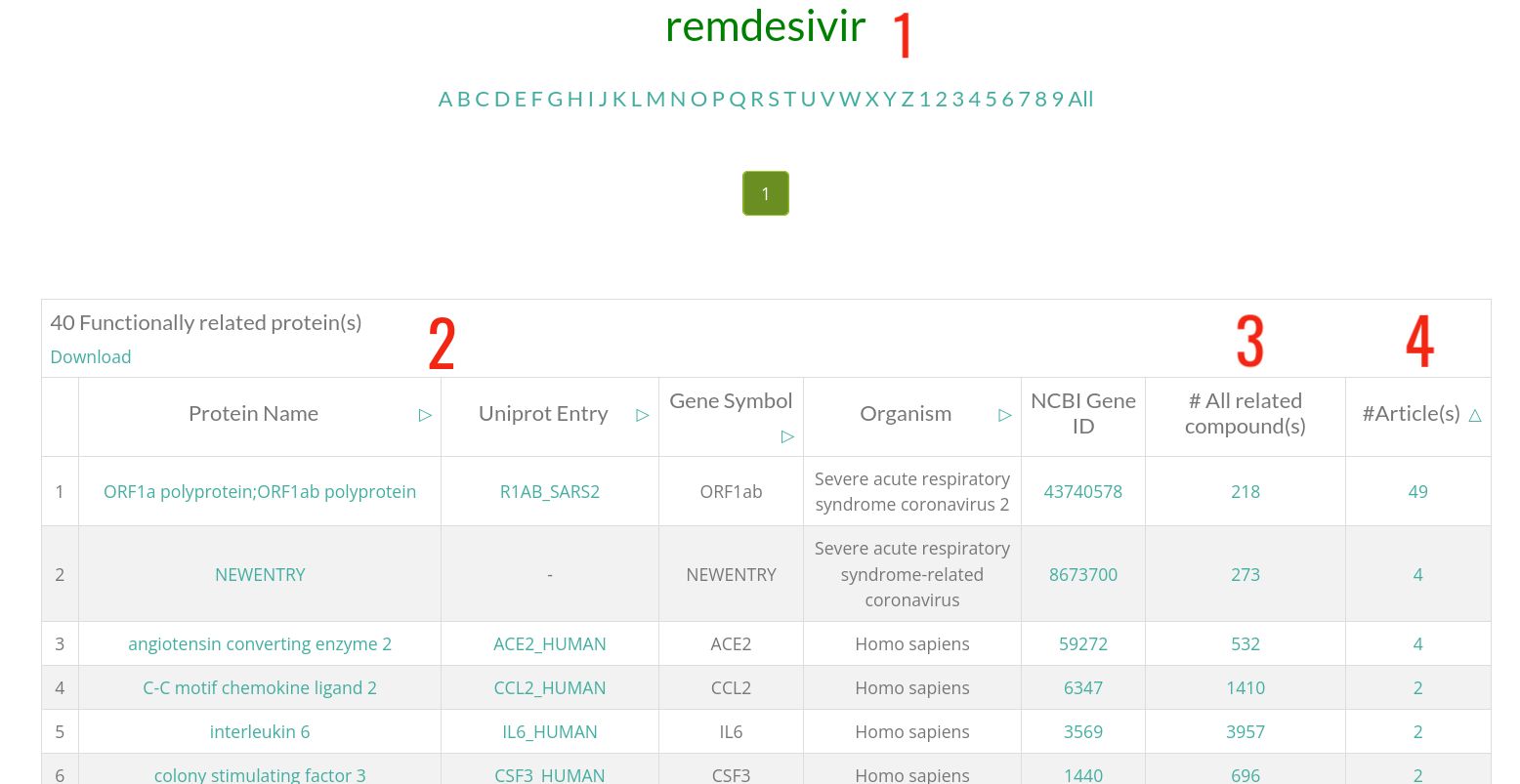
- 1 : Compound name. Click on it to get the related compound card .
- 2 : Information of the protein that is functionally related to the shown compound.
- 3 : Number of total compounds that are functionally related to this protein. Click on the number to see them.
- 4 : Number of abstracts where this compound-protein functional relationship is mentioned. Click on this number to see these articles.
Protein Name
You can search by a specific protein name or gene symbol, this will show the protein with all possible species.
After choosing one of the suggested proteins, all compounds that are functionally related are shown.
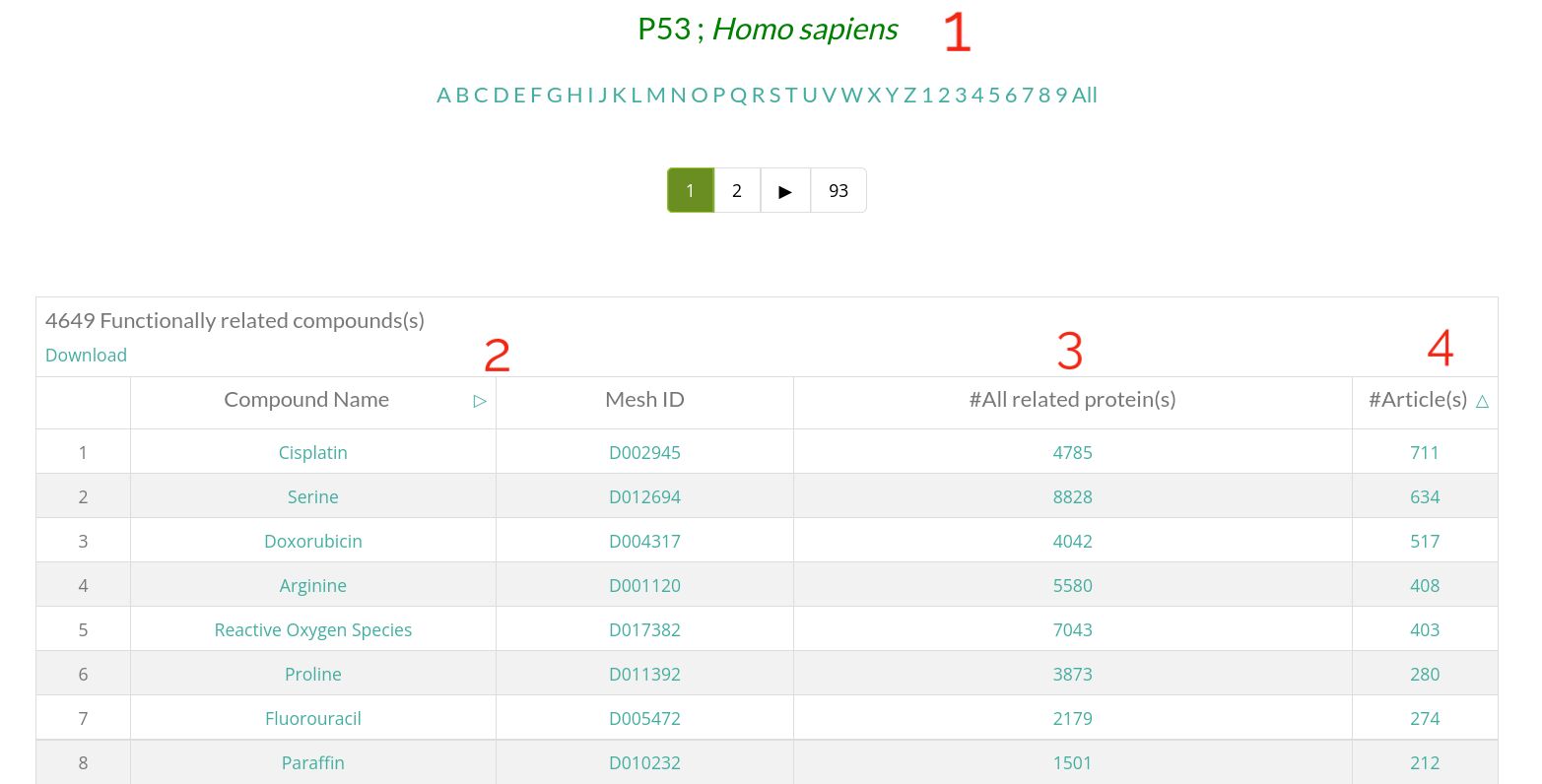
- 1 : Protein Name. Click on it to get the related protein card.
- 2 : Information of the compound that is functionally related to the shown protein.
- 3 : Number of total proteins that are functionally related to this compound. Click on the number to see them.
- 4 : Number of abstracts where this compound-protein functional relationship is mentioned. Click on this number to see these articles.
UniProt Entry Name/UniProt ID
Searching by UniProt entry name/UniProt ID is also possible. As output all compounds that are functionally related are shown.
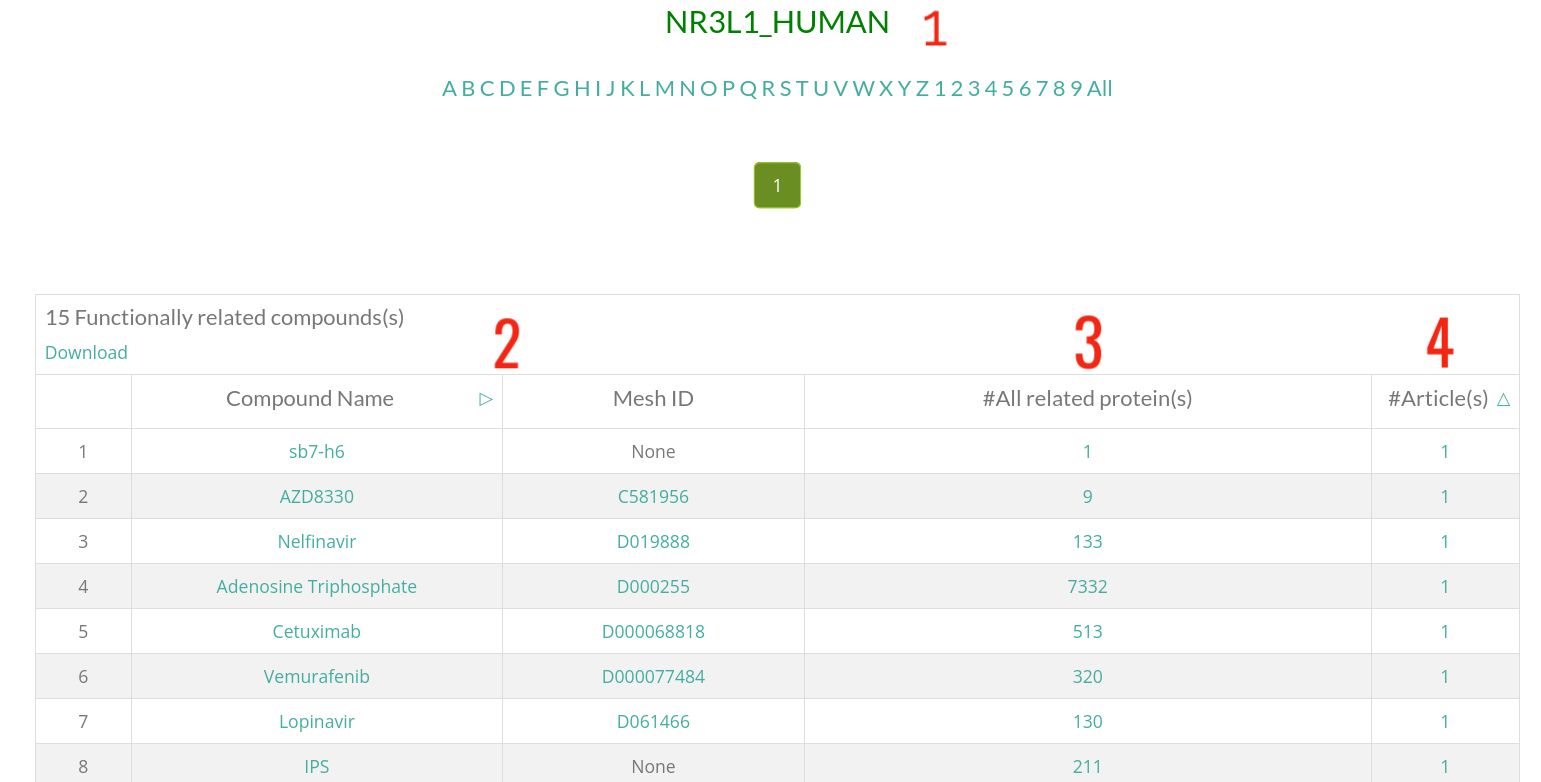
- 1 : UniProt Entry Name/UniProt ID, clicking on it will display the protein card.
- 2 : Information of the compound that is functionally related to the shown protein.
- 3 : Number of total proteins that are functionally related to this compound. Click on the number to see them.
- 4 : Number of abstracts where this compound-protein functional relationship is mentioned. Click on this number to see these articles.
PubMed ID
Here you can look for all compound-protein functional relationships that can be found in specific PubMed abstracts using the PubMed ID.
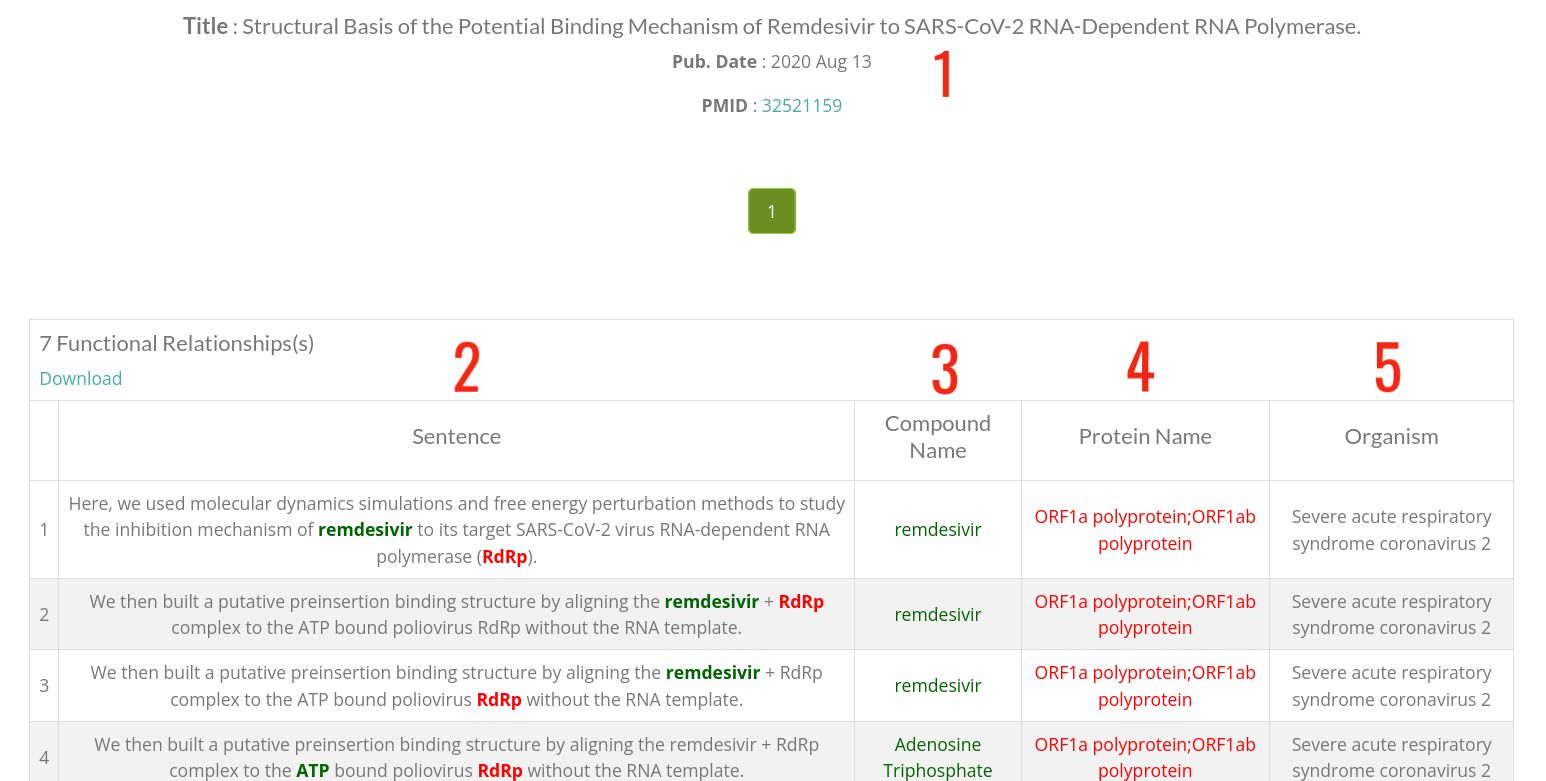
- 1 : Main information of the article.
- 2 : Sentences where the compound-protein functional relationship is mentioned(highlighted both entities).
- 3 : The name of the chemical compound, click to display the compound card).
- 4 : The name of the protein, click to display the protein card).
- 5 : Source organism of the protein.
Advanced Search
You can also find all functional relationships of a comapound-protein pair or an individual entity in a specific period.
Cards
Compound Card
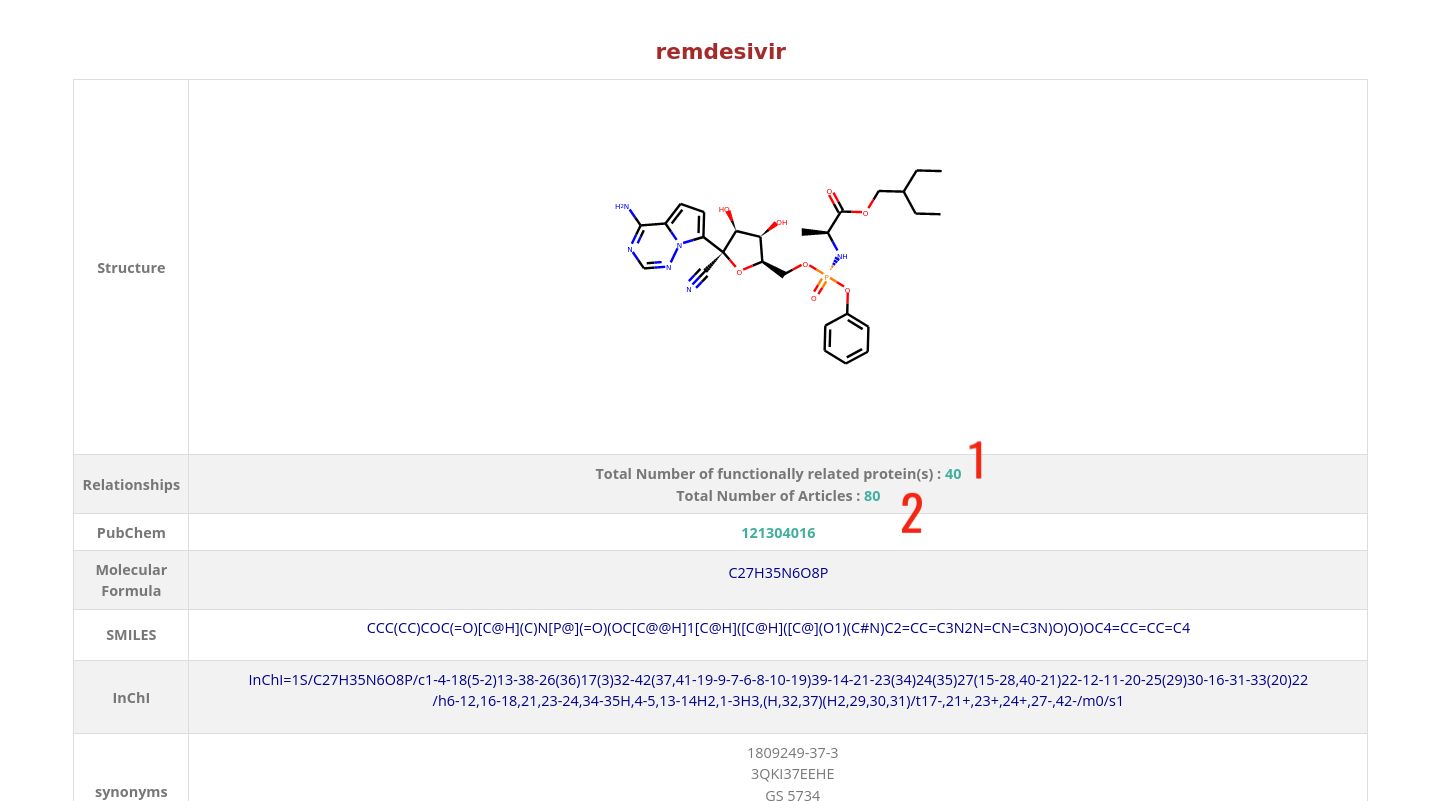
- 1 : Total number of proteins that are functionaly related to this compound.
- 2 : Total number of articles in which relations of this compound are described.
Protein Card
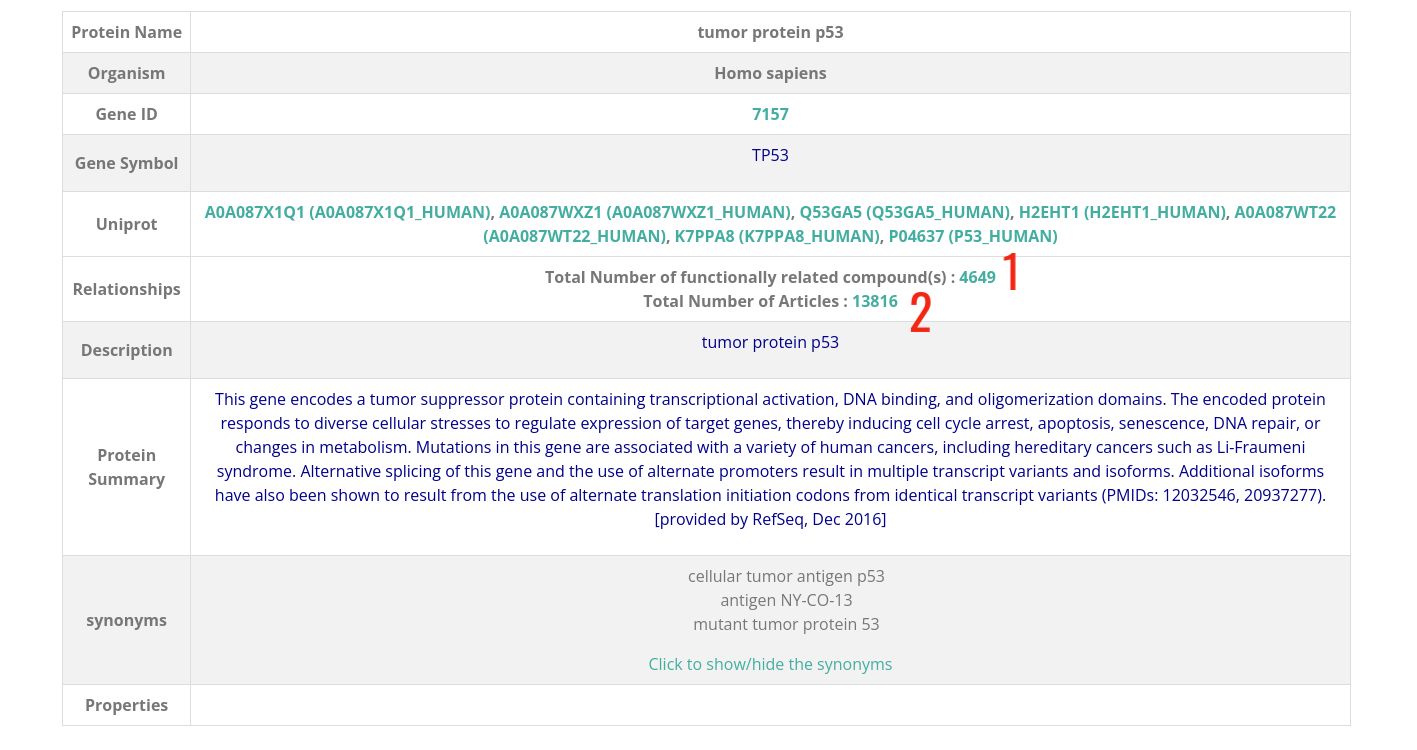
- 1 : Total number of compounds that are related to this protein.
- 2 : Total number of articles in which relations of this protein are described.
Downloads
- Benchmark Dataset: Manual annotation benchmark dataset that used as a training dataset in all techniques that we used to find the functional compound-protein relationships (Please find the related article here).
- 1 : PubMed ID.
- 2 : Text Sentence where the compound-protein functional relationships are found.
- 3 : Entities (compounds and proteins) that are annotated in this sentence.
- 4 : All compound-protein pair combinations.
- 5 : Indicates if there is any functional relationship in this pair(True) or not(False).
- CPRiL-functional compound-protein relationships : The whole functional compound-protein relationships that was extracted from the full PubMed dataset, it's offered in tab-separated format.
The downloaded file is offered in XML format. Here example how this format looks like:

Cite Us
Ammar Qaseem and Stefan Günther
CPRiL: Compound-Protein Relationships in Literature (manuscript in preparation).